Monte Carlo Penalty Selection (MCPeSe) for graphical lasso.
library(huge)
source("mcpese.R")
##########################################################
# Initialize seed number:
#seed = Sys.time()
#seed = as.integer(seed)
#seed = seed %% 100000
seed = 59040
set.seed(seed)
##########################################################
# Graphical model simulation:
p = 200
n = 210
Model = "hub"
HugeData = huge.generator(n=n, d=p, graph=Model)
nlambda = 100
##########################################################
# Compute Glasso solution path:
L = huge(HugeData$data, nlambda=nlambda, method="glasso")
##########################################################
# Run the A-R selection (uniform prior),
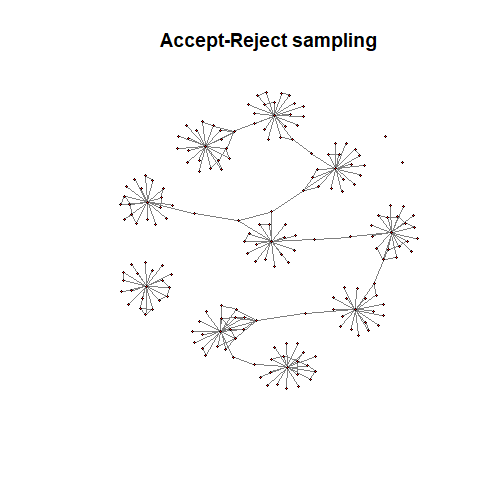
ARSelect = mcpese(L, n=n, M=1000)
names(ARSelect)
rhos = ARSelect$rhos
ARSelect$accept.rate
##########################################################
# Run the M-H selection (uniform prior),
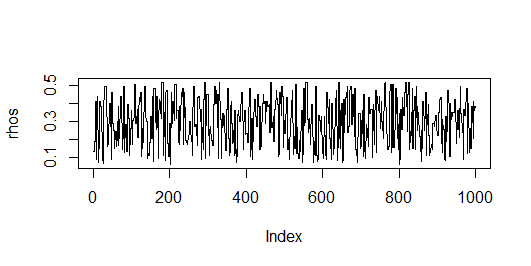
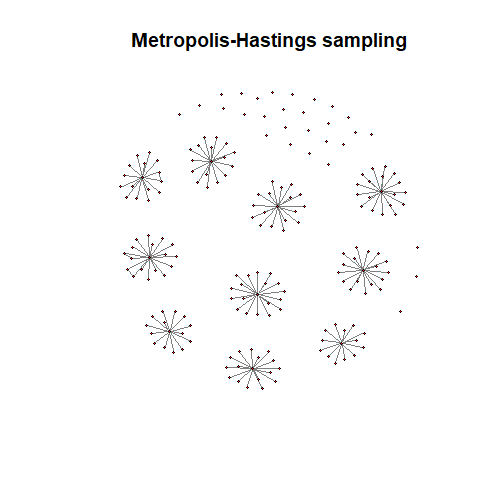
MHSelect = mcpese(L, n=n, nSteps=1000, method="M-H")
names(MHSelect)
rhos = MHSelect$rhos
MHSelect$accept.rate
plot(rhos, type="l")
########################################################### Either use the mean of rho values...
mean(rhos)
#ThetaARSelect = huge(Y, lambda=mean(rhos), method="glasso")
#ThetaAR = as.matrix(ThetaARSelect$icov[[1]])
# ... or pick the smallest tuning parameter value from the solution path
# which is larger or equal to the mean value:
optARrhoIndx = ARSelect$opt.index # This is sup{i : rho[i] >= mean(rhos)}
optMHrhoIndx = MHSelect$opt.index
huge.plot(HugeData$theta)
title("Ground truth")
##########################################################huge.plot(L$path[[optARrhoIndx]])
title("Accept-Reject sampling")
##########################################################huge.plot(L$path[[optMHrhoIndx]])
title("Metropolis-Hastings sampling")The MCPeSe method is described in:
Kuismin and Sillanpaa (2020). MCPeSe: Monte Carlo penalty selection for graphical lasso, Bioinformatics, https://doi.org/10.1093/bioinformatics/btaa734.
File "CodeCollection.zip" is a collection of scripts used to prepare the material in this paper.